Part 2 Data manipulation with R
2.1 Get Started
Multiple choices cases
- R base
- Tidyverse
- datatable
2.2 Data wrangling with R base
2.2.1 Load data
2.2.1.1 Load data from an embedded data set:
LakeHuron <- LakeHuron: loads explicitly the Lake Huron dataset- Look at the Environnement panel
- type
LakeHuron
This can be useful to reproduce documentation examples but it is not your data. You need to load data from the outside.
2.2.1.2 Load data from a csv file
"data/worldbank_df.csv" is the path to the file, from the working directory.
Note that, even on Microsoft Windows, path notation use slashes / to separate directories.
sep* and header are options to set accordingly to your file.
This would have work too:
2.2.1.3 What’s the difference between LakeHuron and worldbank ?
Tips :
- Look into the Environnement panel
- use the class function
`LakeHuron ̀is a time series, there is just one column with the water height. But the start and ending years are indicated
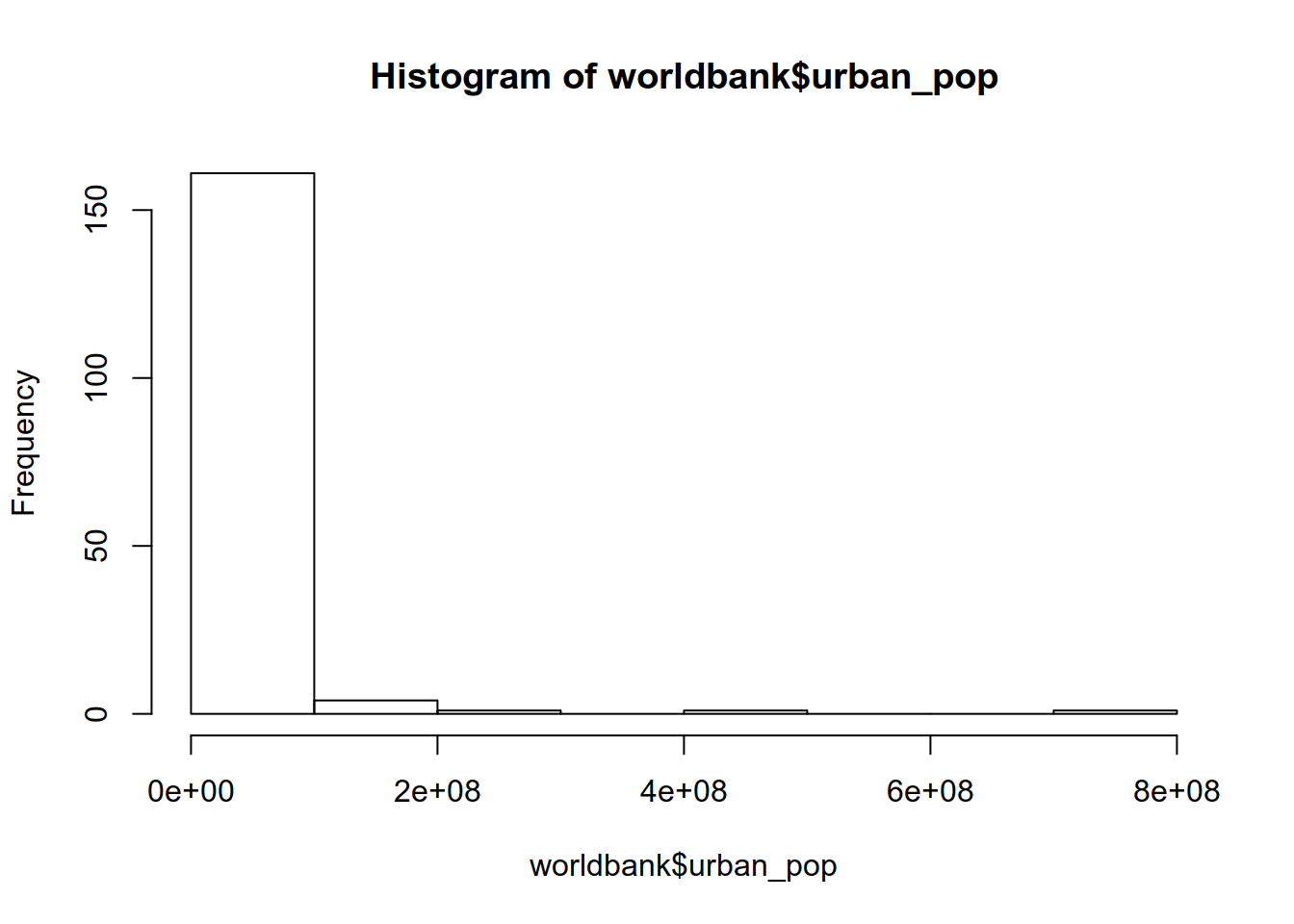
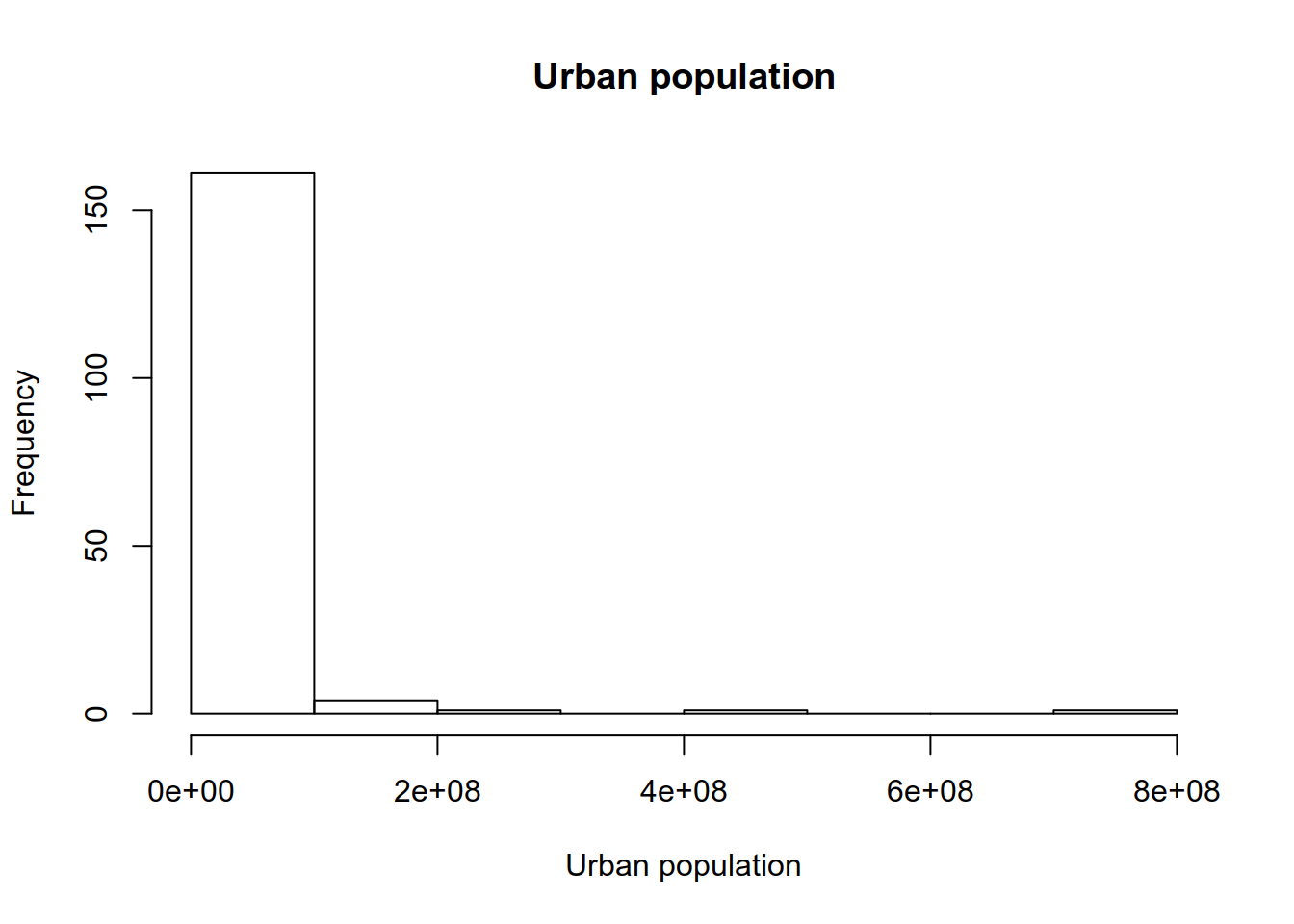
̀worldbank_df ̀ is a dataframe with several columns (name, urban pop)
2.2.2 Get some insight about the data
2.2.2.1 Dataset dimensions
length(): to get the length of a vector
## [1] 98There is 98 records in the LakeHuron dataset.
nrow()̀̀: number of rows of a dataset
## [1] 177There is 177 rows in the worldbank dataset.
dim(): dimensions (number of rows and columns) of a dataset
## [1] 177 7There is 177 rows and 7 columns in the dataset.
2.2.2.2 Descriptives statistics
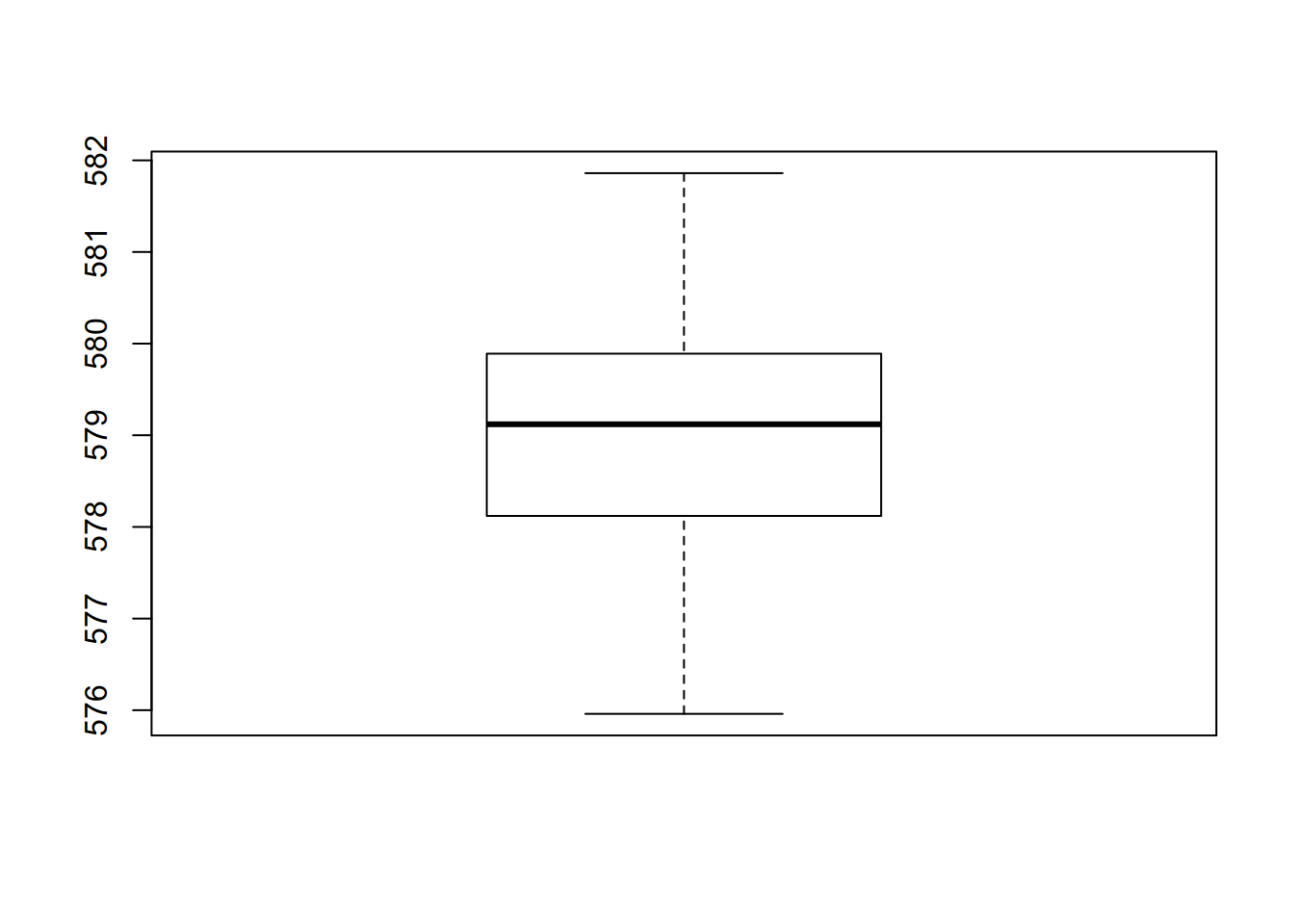
- Mean
## [1] 579.0041- Median
## [1] 579.12- Minimum
## [1] 575.96- Maximum
## [1] 581.86- Range
## [1] 575.96 581.86- Quantiles
## 0% 25% 50% 75% 100%
## 575.960 578.135 579.120 579.875 581.860- Variance
## [1] 1.737911- Standard deviation
## [1] 1.318299- Summary
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 576.0 578.1 579.1 579.0 579.9 581.9The na.rm = TRUE option remove the Non Available data before computation in
order to avoid taking it in account as zeros.
2.2.2.3 Work with columns
2.2.2.3.1 How to get the column names ?
## [1] "name" "iso_a2" "HDI" "urban_pop"
## [5] "unemployment" "pop_growth" "literacy"##### Get the content of a column
- as an object
## [1] Afghanistan Angola Albania
## [4] United Arab Emirates Argentina Armenia
## 177 Levels: Afghanistan Albania Algeria Angola Antarctica ... ZimbabweThe head() function display only the 6 first rows, what does the tail() function ?
- By column name
## name
## 1 Afghanistan
## 2 Angola
## 3 Albania
## 4 United Arab Emirates
## 5 Argentina
## 6 Armenia- By column order
## [1] Afghanistan Angola Albania
## [4] United Arab Emirates Argentina Armenia
## 177 Levels: Afghanistan Albania Algeria Angola Antarctica ... ZimbabweIn brackets first digits are the row numbers, after the comma it is the column numbers.
2.2.2.3.2 Slicing
Syntax : Numbers between brackets: [2:3]
- Create an example vector
a
## [1] 1 2 3 4 5 6 7 8 9 10- Slice the
avector
## [1] 2 3 4 5- Slicing works with columns
Numbers between brackets: [<rows> , <columns>]
- Only the columns 2 and 3
## [,1] [,2]
## [1,] 4 7
## [2,] 5 8
## [3,] 6 9- Just the first row of the columns 2 and 3
## [1] 6 9What is that code doing ?
## [1] Afghanistan Angola Albania
## [4] United Arab Emirates Argentina Armenia
## 177 Levels: Afghanistan Albania Algeria Angola Antarctica ... Zimbabwe2.2.2.3.2.1 Exercice
Try to get the 10 first rows and the 3 first columns of the worldbank dataset.
## name iso_a2 HDI
## 1 Afghanistan AF NA
## 2 Angola AO 0.504
## 3 Albania AL NA
## 4 United Arab Emirates AE NA
## 5 Argentina AR NA
## 6 Armenia AM NA
## 7 Antarctica AQ NA
## 8 French Southern and Antarctic Lands TF NA
## 9 Australia AU NA
## 10 Austria AT NA## name iso_a2 HDI
## 1 Afghanistan AF NA
## 2 Angola AO 0.504
## 3 Albania AL NA
## 4 United Arab Emirates AE NA
## 5 Argentina AR NA
## 6 Armenia AM NA
## 7 Antarctica AQ NA
## 8 French Southern and Antarctic Lands TF NA
## 9 Australia AU NA
## 10 Austria AT NAFor more precision on slicing, use vectors:
## name urban_pop
## 1 Afghanistan 8609463
## 3 Albania 1629715
## 5 Argentina 39372787## name urban_pop
## 1 Afghanistan 8609463
## 3 Albania 1629715
## 5 Argentina 393727872.3 Data wrangling with the Tidyverse
2.3.1 First load the tidyverse package
## ── Attaching packages ────────────────────────────────── tidyverse 1.3.0 ──## ✔ ggplot2 3.2.1 ✔ purrr 0.3.3
## ✔ tibble 2.1.3 ✔ dplyr 0.8.3
## ✔ tidyr 1.0.0 ✔ stringr 1.4.0
## ✔ readr 1.3.1 ✔ forcats 0.4.0## ── Conflicts ───────────────────────────────────── tidyverse_conflicts() ──
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::lag() masks stats::lag()Then ask questions
- most useful tydiverse package for data manipulation is dplyr
- each action is a verb
- filter – select a subset of the rows of a data frame
- arrange – works similarly to filter, except that instead of filtering or selecting rows, it reorders them
- select – select columns of a data frame
- mutate – add new columns to a data frame that are functions of existing columns
- summarize – summarize values
- group_by – describe how to break a data frame into groups of rows
- you can pipe successive actions with a
%>% - Tibbles: dataframes++
2.3.2 Dataset
We will use the gapminder dataset:
## # A tibble: 6 x 6
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Afghanistan Asia 1952 28.8 8425333 779.
## 2 Afghanistan Asia 1957 30.3 9240934 821.
## 3 Afghanistan Asia 1962 32.0 10267083 853.
## 4 Afghanistan Asia 1967 34.0 11537966 836.
## 5 Afghanistan Asia 1972 36.1 13079460 740.
## 6 Afghanistan Asia 1977 38.4 14880372 786.The gapminder data frames include six variables, (Gapminder.org
documentation page):
| variable | meaning |
|---|---|
| country | |
| continent | |
| year | |
| lifeExp | life expectancy at birth |
| pop | total population |
| gdpPercap | per-capita GDP |
Per-capita GDP (Gross domestic product) is given in units of international dollars, “a hypothetical unit of currency that has the same purchasing power parity that the U.S. dollar had in the United States at a given point in time” – 2005, in this case.
gapminder: 12 rows for each country (1952, 1955, …, 2007).
2.3.3 select()
Select one or more column by name. Let’s keep only the year, country and
gdpPercap columns.
## # A tibble: 1,704 x 3
## year country gdpPercap
## <int> <fct> <dbl>
## 1 1952 Afghanistan 779.
## 2 1957 Afghanistan 821.
## 3 1962 Afghanistan 853.
## 4 1967 Afghanistan 836.
## 5 1972 Afghanistan 740.
## 6 1977 Afghanistan 786.
## 7 1982 Afghanistan 978.
## 8 1987 Afghanistan 852.
## 9 1992 Afghanistan 649.
## 10 1997 Afghanistan 635.
## # … with 1,694 more rows2.3.4 filter()
Filter rows matching a condition (TRUE or FALSE test). We will keep only records in Europe.
year_country_gdp_euro <- gapminder %>%
filter(continent == "Europe") %>%
select(year, country, gdpPercap)
year_country_gdp_euro## # A tibble: 360 x 3
## year country gdpPercap
## <int> <fct> <dbl>
## 1 1952 Albania 1601.
## 2 1957 Albania 1942.
## 3 1962 Albania 2313.
## 4 1967 Albania 2760.
## 5 1972 Albania 3313.
## 6 1977 Albania 3533.
## 7 1982 Albania 3631.
## 8 1987 Albania 3739.
## 9 1992 Albania 2497.
## 10 1997 Albania 3193.
## # … with 350 more rowsExercice 1
Produce a dataframe that has the African values for lifeExp, country and year, but not for other Continents
## # A tibble: 624 x 3
## year country lifeExp
## <int> <fct> <dbl>
## 1 1952 Algeria 43.1
## 2 1957 Algeria 45.7
## 3 1962 Algeria 48.3
## 4 1967 Algeria 51.4
## 5 1972 Algeria 54.5
## 6 1977 Algeria 58.0
## 7 1982 Algeria 61.4
## 8 1987 Algeria 65.8
## 9 1992 Algeria 67.7
## 10 1997 Algeria 69.2
## # … with 614 more rows2.3.5 mutate()
mutate() allows to add new columns to a dataset. transmute() use the same syntax but only keep the new columns.
## # A tibble: 1,704 x 7
## country continent year lifeExp pop gdpPercap total_gdp
## <fct> <fct> <int> <dbl> <int> <dbl> <dbl>
## 1 Afghanistan Asia 1952 28.8 8425333 779. 6567086330.
## 2 Afghanistan Asia 1957 30.3 9240934 821. 7585448670.
## 3 Afghanistan Asia 1962 32.0 10267083 853. 8758855797.
## 4 Afghanistan Asia 1967 34.0 11537966 836. 9648014150.
## 5 Afghanistan Asia 1972 36.1 13079460 740. 9678553274.
## 6 Afghanistan Asia 1977 38.4 14880372 786. 11697659231.
## 7 Afghanistan Asia 1982 39.9 12881816 978. 12598563401.
## 8 Afghanistan Asia 1987 40.8 13867957 852. 11820990309.
## 9 Afghanistan Asia 1992 41.7 16317921 649. 10595901589.
## 10 Afghanistan Asia 1997 41.8 22227415 635. 14121995875.
## # … with 1,694 more rows- Change mutate() by transmute() and compare the results.
- How to keep the
yearandcountrycolumn ?
2.3.6 group_by()
Group data by a variable.
## Classes 'tbl_df', 'tbl' and 'data.frame': 1704 obs. of 6 variables:
## $ country : Factor w/ 142 levels "Afghanistan",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ continent: Factor w/ 5 levels "Africa","Americas",..: 3 3 3 3 3 3 3 3 3 3 ...
## $ year : int 1952 1957 1962 1967 1972 1977 1982 1987 1992 1997 ...
## $ lifeExp : num 28.8 30.3 32 34 36.1 ...
## $ pop : int 8425333 9240934 10267083 11537966 13079460 14880372 12881816 13867957 16317921 22227415 ...
## $ gdpPercap: num 779 821 853 836 740 ...## Classes 'tbl_df', 'tbl' and 'data.frame': 1704 obs. of 6 variables:
## $ country : Factor w/ 142 levels "Afghanistan",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ continent: Factor w/ 5 levels "Africa","Americas",..: 3 3 3 3 3 3 3 3 3 3 ...
## $ year : int 1952 1957 1962 1967 1972 1977 1982 1987 1992 1997 ...
## $ lifeExp : num 28.8 30.3 32 34 36.1 ...
## $ pop : int 8425333 9240934 10267083 11537966 13079460 14880372 12881816 13867957 16317921 22227415 ...
## $ gdpPercap: num 779 821 853 836 740 ...## Classes 'grouped_df', 'tbl_df', 'tbl' and 'data.frame': 1704 obs. of 6 variables:
## $ country : Factor w/ 142 levels "Afghanistan",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ continent: Factor w/ 5 levels "Africa","Americas",..: 3 3 3 3 3 3 3 3 3 3 ...
## $ year : int 1952 1957 1962 1967 1972 1977 1982 1987 1992 1997 ...
## $ lifeExp : num 28.8 30.3 32 34 36.1 ...
## $ pop : int 8425333 9240934 10267083 11537966 13079460 14880372 12881816 13867957 16317921 22227415 ...
## $ gdpPercap: num 779 821 853 836 740 ...
## - attr(*, "groups")=Classes 'tbl_df', 'tbl' and 'data.frame': 5 obs. of 2 variables:
## ..$ continent: Factor w/ 5 levels "Africa","Americas",..: 1 2 3 4 5
## ..$ .rows :List of 5
## .. ..$ : int 25 26 27 28 29 30 31 32 33 34 ...
## .. ..$ : int 49 50 51 52 53 54 55 56 57 58 ...
## .. ..$ : int 1 2 3 4 5 6 7 8 9 10 ...
## .. ..$ : int 13 14 15 16 17 18 19 20 21 22 ...
## .. ..$ : int 61 62 63 64 65 66 67 68 69 70 ...
## ..- attr(*, ".drop")= logi TRUEIf you look at the grouped dataframe, you can see that group_by created
new smaller dataframes (one by continent) along side the original one.
All following calculations within the summarize() function will be made on those separated dataframes.
2.3.7 summarize()
summarize() aggregates the grouped data with a function (sum, mean, custom, etc.).
gdp_bycontinents <- gapminder %>%
group_by(continent) %>%
summarize(
mean_gdpPercap=mean(gdpPercap)
)
gdp_bycontinents## # A tibble: 5 x 2
## continent mean_gdpPercap
## <fct> <dbl>
## 1 Africa 2194.
## 2 Americas 7136.
## 3 Asia 7902.
## 4 Europe 14469.
## 5 Oceania 18622.]
Exercice 2
Calculate mean and median life expectancy in South Africa and Ireland
Steps
- Select the columns
- Filter the countries
- summarize the data in 2 new variables
Expected output
## # A tibble: 1 x 2
## AverageLife MedianLife
## <dbl> <dbl>
## 1 63.5 64.4Exercice 3
Calculate the gap between a mean life expectancy in a country and the world mean life expectancy.
Steps
- Select the columns
- Create a new variable
lifeExp_gap - Group data by
country - Summarize by creating 2 new variables
country_mean_life_expandcountry_mean_life_exp_gap
Expected output
## # A tibble: 142 x 3
## country country_mean_life_exp country_mean_life_exp_gap
## <fct> <dbl> <dbl>
## 1 Afghanistan 37.5 -22.0
## 2 Albania 68.4 8.96
## 3 Algeria 59.0 -0.444
## 4 Angola 37.9 -21.6
## 5 Argentina 69.1 9.59
## 6 Australia 74.7 15.2
## 7 Austria 73.1 13.6
## 8 Bahrain 65.6 6.13
## 9 Bangladesh 49.8 -9.64
## 10 Belgium 73.6 14.2
## # … with 132 more rowsHint
2.3.7.1 Sort data
If needed you can arrange rows by life expectancy gaps.
- Ascending order
- Descending order
mean_lifeExp_gap <- mean_lifeExp_gap %>%
arrange(desc(country_mean_life_exp_gap))
head(mean_lifeExp_gap)## # A tibble: 6 x 3
## country country_mean_life_exp country_mean_life_exp_gap
## <fct> <dbl> <dbl>
## 1 Sierra Leone 36.8 -22.7
## 2 Afghanistan 37.5 -22.0
## 3 Angola 37.9 -21.6
## 4 Guinea-Bissau 39.2 -20.3
## 5 Mozambique 40.4 -19.1
## 6 Somalia 41.0 -18.5## # A tibble: 6 x 3
## country country_mean_life_exp country_mean_life_exp_gap
## <fct> <dbl> <dbl>
## 1 Iceland 76.5 17.0
## 2 Sweden 76.2 16.7
## 3 Norway 75.8 16.4
## 4 Netherlands 75.6 16.2
## 5 Switzerland 75.6 16.1
## 6 Canada 74.9 15.42.3.8 Commands order
Even not mandatory, it is recommended to use that order:
filterthe rowsselectthe columnsmutateif neededgroup byif neededsummarizeif needed
filterandselectfirst to reduce the size of the dataset to handle.filterfirst to avoid issues by not selecting columns that are used to filter.
Now let’s see how to plot with ggplot2